Preliminary Information
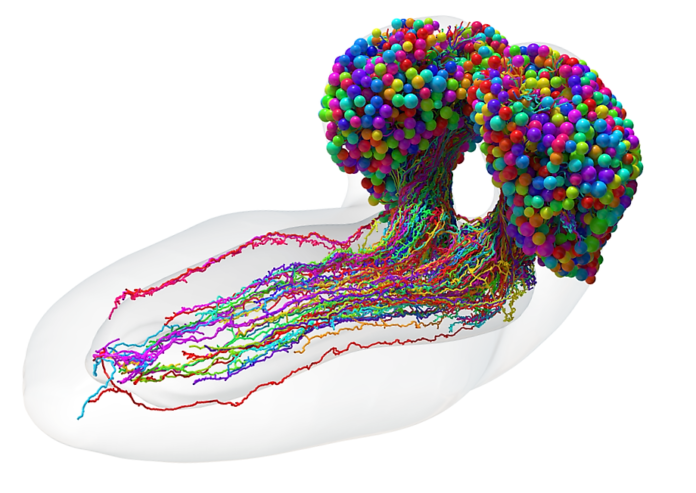
Behold, as the intrepid scientists have succeeded in developing the very first complete connectome of a six-hour-old female Drosophila melanogaster larval brain! A vast and intricate web of more than half a million neural connections comprise the wiring map, which features a tremendous amount of feedback loops and layered pathways to facilitate optimal support for the brain’s learning capabilities.
Intricate Details of the Connectome
Spanning all 3,016 neurons and 548,000 synapses in the central nervous system of the larval fruit fly, the connectome encompasses everything from the axons that deliver sensory input to the brain to the dendrites on neurons that send out brain signals to control movement. The MRC Laboratory of Molecular Biology at the University of Cambridge, UK spearheaded the research effort, and the outcomes are truly remarkable.
Elucidation of Brain Architecture and Behaviors
Fruit fly larvae are more complex than previously researched organisms and have more complex behaviors such as moving toward the smell of food and away from light. Such actions require intricate brain structures capable of converting sensory information into movement. The new wiring map traces how the sensory input traverses to the motor output with the aid of both short and long pathways. In addition, the two-way recurrent connections within neurons play an essential role in learning and information processing.
Significance of the Connectome
Understanding the fundamental principles of information processing in the brain is critical, and this work is instrumental in achieving this goal. Researchers have generated larger yet incomplete connectomes for more complex brains, such as a map of more than 20 million connections among 25,000 neurons in the adult fruit fly. This new map can serve as a standard for comparison with connectomes of larvae with genetic mutations or atypical environmental conditions.
Future Research Directions
The creation of the new wiring map took five years, including more than a year merely to image the brain slices. However, new electron microscopes and software can trace neurons much more efficiently, and scientists plan to take advantage of these developments. Optogenetics is expected to be used to generate activity maps of the larval brain to understand how the connections operate during different behaviors. This could assist in understanding gene mutations related to autism, as many of the genes are essential for synapses to function.
Implications for Neuroscientific Research
The development of a complete wiring map of the larval fruit fly brain has significant implications for neuroscientific research. This new map provides researchers with a detailed blueprint of the neural connections that underlie complex behaviors, opening up new avenues for understanding the underlying mechanisms of learning and memory. Moreover, the map can serve as a valuable tool for studying the effects of genetic mutations or environmental factors on brain development and function.
Conclusion
In summary, the creation of a complete wiring map of the larval fruit fly brain is a significant achievement in the field of neuroscience. The map provides a detailed account of the neural connections that support complex behaviors, offering insights into the fundamental principles of information processing in the brain. Moreover, the map can serve as a valuable resource for future research aimed at understanding the effects of genetic mutations or environmental factors on brain development and function. With the advent of new imaging technologies and analytical tools, the future of neuroscience research looks bright, and we can expect to make further significant strides in understanding the brain’s complexities.
Google News | Telegram