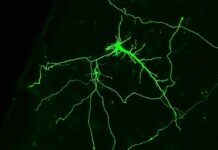
A Paradigm Shift in Single-Cell Sequencing
With a groundbreaking combination of long-read technology and single-cell sequencing, researchers have achieved an unparalleled understanding of mutational differences between individual cell genomes. Single-cell sequencing has long been hampered by the challenge of capturing complex repetitive regions of DNA, as previous methods only allowed for the reading of the genome in short sections. Long-read technology has the potential to overcome this problem, but the requirement of several micrograms of genetic material posed an insurmountable obstacle as a single cell contains only six picograms of genetic material. However, the use of innovative DNA amplification techniques, such as droplet-based multiple displacement amplification, has enabled scientists to perform long-read sequencing from single cells, unlocking a new realm of genetic analysis.
The Significance of Long-Read Sequencing and DNA Amplification Techniques
For the past decade, long-read sequencing has revolutionized the scientific community’s understanding of the “dark regions” of the genome that are inaccessible to short-read technologies. However, the amount of DNA required for the process was simply too high for single-cell sequencing. The droplet-based amplification technique, by preventing amplification bias, has successfully addressed this hurdle. The technique’s success lies in its ability to amplify a tiny amount of DNA into a large quantity without introducing bias, enabling long-read sequencing from single cells.
Unlocking the Potential of Single-Cell Long-Read Sequencing
Improved single-cell DNA sequencing has the potential to unlock insights into the mutations that make tumors more aggressive by identifying sub-clonal mutations. Furthermore, the technique could aid in the study of somatic mutations that are believed to be associated with neurological conditions such as Tourette’s, schizophrenia, and autism. Long-read sequencing of single cells could also help identify genetic variants in speech disorders, facilitating early diagnosis and intervention.
Limitations and Future Prospects
While the droplet-based amplification technique has reduced amplification bias, it is not without limitations, such as errors and chimeras, where non-neighboring parts of the genome are combined. Researchers are working tirelessly to optimize the technique to minimize errors during amplification. The study serves as a proof-of-principle that demonstrates the potential of long-read sequencing at the single-cell level. The researchers’ plan to refine the technique to enable even more in-depth studies of single cells in the future.
Conclusion
The breakthrough ability to perform single-cell long-read sequencing opens up a new era in genetic analysis. Researchers can now combine innovative approaches to sequence the full spectrum of mutational differences between individual cells’ genomes. The improved technique holds immense promise in identifying sub-clonal mutations in cancer, studying the impact of somatic mutations in neurological conditions, and identifying genetic variants in speech disorders. Despite some limitations, researchers believe that this proof-of-principle study represents a crucial step in the development of long-read sequencing at the single-cell level, promising exciting prospects for future genetic analysis.
Google News | Telegram